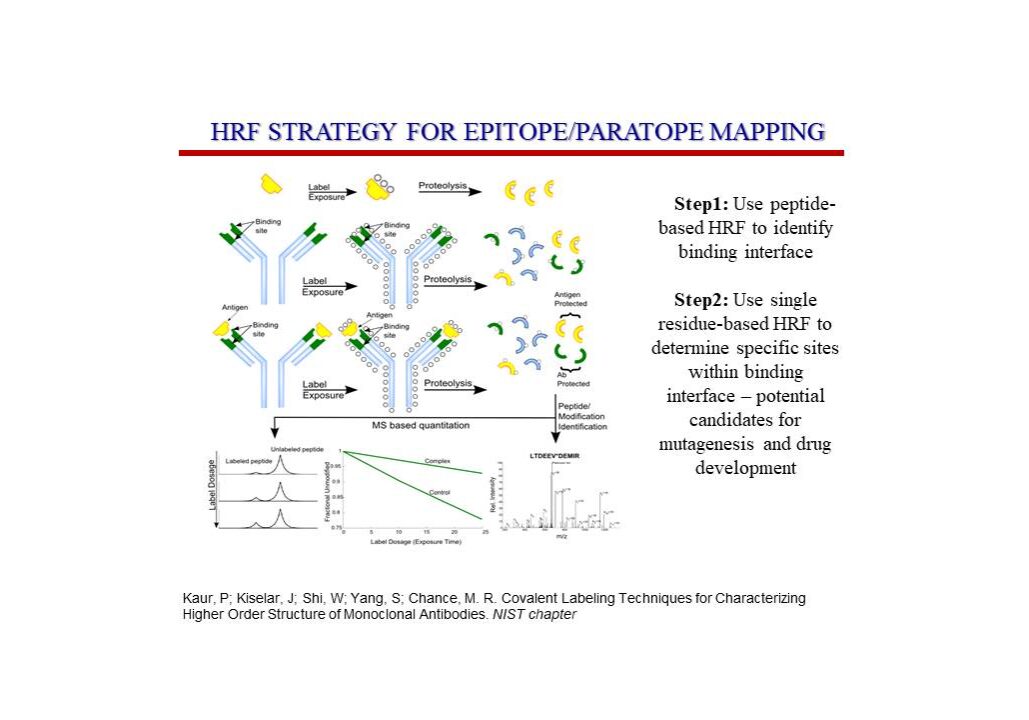
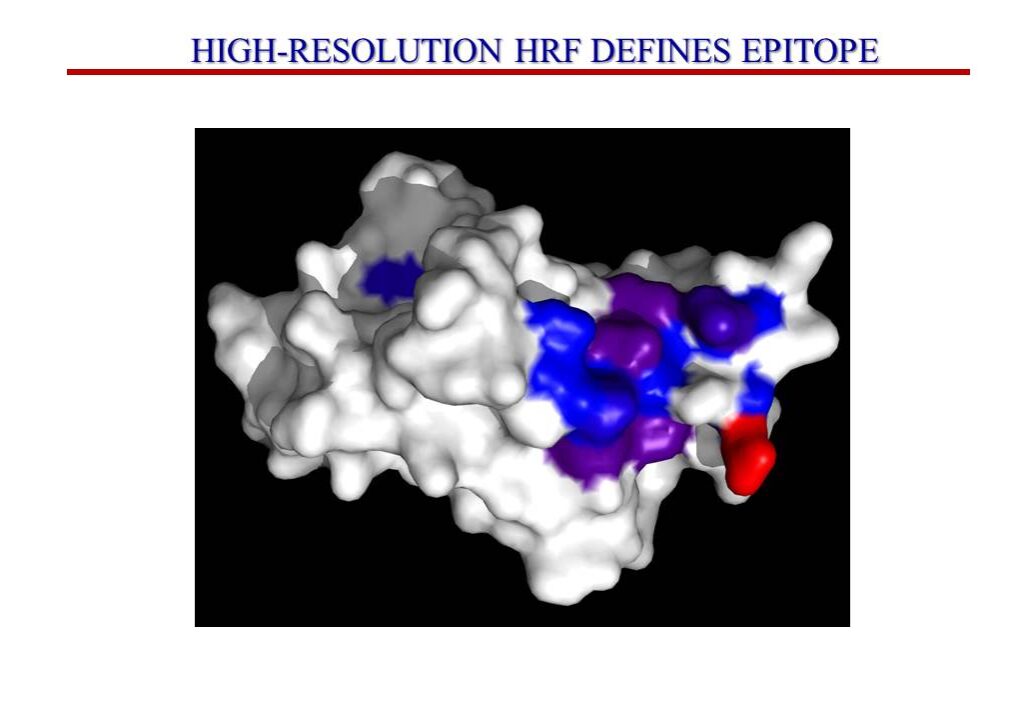
Epitope Mapping Service
Neo offers covalent labeling (CL) and mass spectrometry (MS) services to industry professionals to characterize therapeutic proteins. These services precisely define epitopes for antigen targets and provide structural characterization of therapeutic proteins suitable for biosimilarity comparisons or any relevant structural definition of the product. Specifically, CL-MS (often called protein footprinting) provides a high-resolution and sensitive readout of the side chain environment (e.g., solvent accessibility) for virtually all side chains of a protein of interest. Changes in solvent accessibility of side chains due to ligand binding (e.g., epitope mapping), sequence or process variation, or changes in solvent are easily read out by the technology and are immediately structurally interpretable.
A variety of labeling techniques such as OH radicals (generated using x-ray radiolysis, Fenton chemistry, or photolysis of peroxide), carbene labeling, carbodiimide, and diethylpyrocarbonate labeling reagents are commonly used in structural mass spectrometry experiments. Labeling can be performed with either less-specific labels, such as hydroxyl radicals that can label most side chains, or more specific labels using chemical reagents that target particular residues.
Workflow of a Typical Hydroxyl Radical-Based Footprinting Experiment
There are a variety of chemistries and labeling methods available. Ask us which might be best for you.