Protein Footprinting – Introduction to Covalent Labeling Technology
Protein footprinting, also called covalent labeling, was initially pioneered by Galas and Schmitz in 1978 to characterize the sites of DNA-protein interaction. The principle behind covalent labeling approaches is that the solvent-accessible side chains of the protein gain stable covalent modifications upon exposure to specific labeling reagents. The ligand-bound form of the given protein experiences significant reduction in the solvent accessibility at the tight ligand-binding site. Thus, when the protein-ligand complex is exposed to the labeling reaction, specific regions of the protein are protected and unable to be modified by the labeling reagents. The differences in the extent of modification as a function of ligand binding or unfolding provide insights into underlying structural changes.
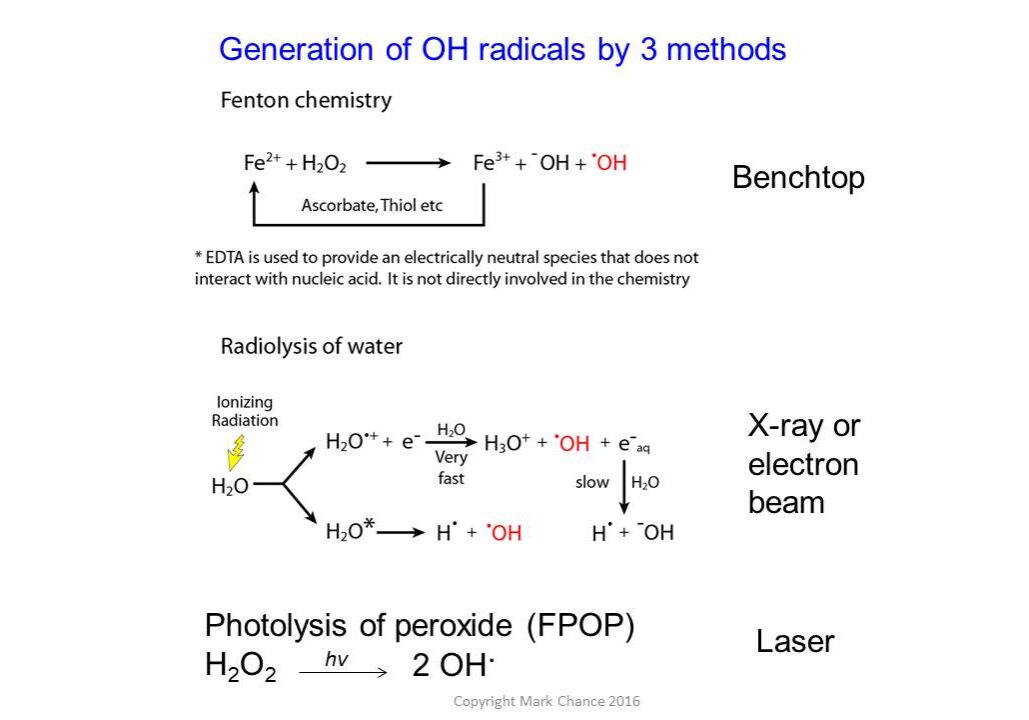
A variety of labeling techniques such as OH radicals (generated using x-ray radiolysis, Fenton chemistry, or photolysis of peroxide), carbene labeling, carbodiimide, and diethylpyrocarbonate labeling reagents are commonly used in structural mass spectrometry experiments. Labeling can be performed with either less-specific labels, such as hydroxyl radicals that can label most side chains, or more specific labels using chemical reagents that target particular residues.
Workflow of a Typical Hydroxyl Radical-Based Footprinting Experiment
There are a variety of chemistries and labeling methods available. Ask us which might be best for you.
The protein solution is exposed to hydroxyl radicals, leading to covalent oxidative modifications on the surface-accessible residues. The exposure experiment is carried out for each individual protein and for the protein complex. The exposed samples undergo proteolysis followed by high-pressure liquid chromatography separation coupled with mass spectrometry analysis. The stable side chain oxidative modifications result in specific mass shifts, which can be identified and localized using tandem mass spectrometry methods. The selected ion chromatograms (SIC) are extracted for the unoxidized and all oxidized forms of the peptide ion (with particular m/z). Integration of peak areas of each isoform in SIC provides the respective total ion abundance. These abundance values are used to characterize reaction kinetics in the form of dose response plots (bottom panel), which measure the “loss of intact peptide” as a function of the hydroxyl radical exposure. The solvent “protected” regions in the complex experience gradual oxidation reaction, as opposed to the unliganded form of the protein. Differences in the rate of oxidation serve to highlight the location of the binding site.
What do footprinting experiments tell you?
Footprinting experiments provide:
- An understanding of macromolecular folding and interactions in solution, even for very large assemblies.
- A measure of solvent accessibility of local sites (tertiary contacts) within a macromolecule.
- An understanding of all solvent-accessible side chain residues for hydroxyl radical-mediated footprinting of proteins and other reagents as needed.
- The ability to probe dynamics on millisecond timescales or equilibrium structures. Requires femtomoles to picomoles of material.
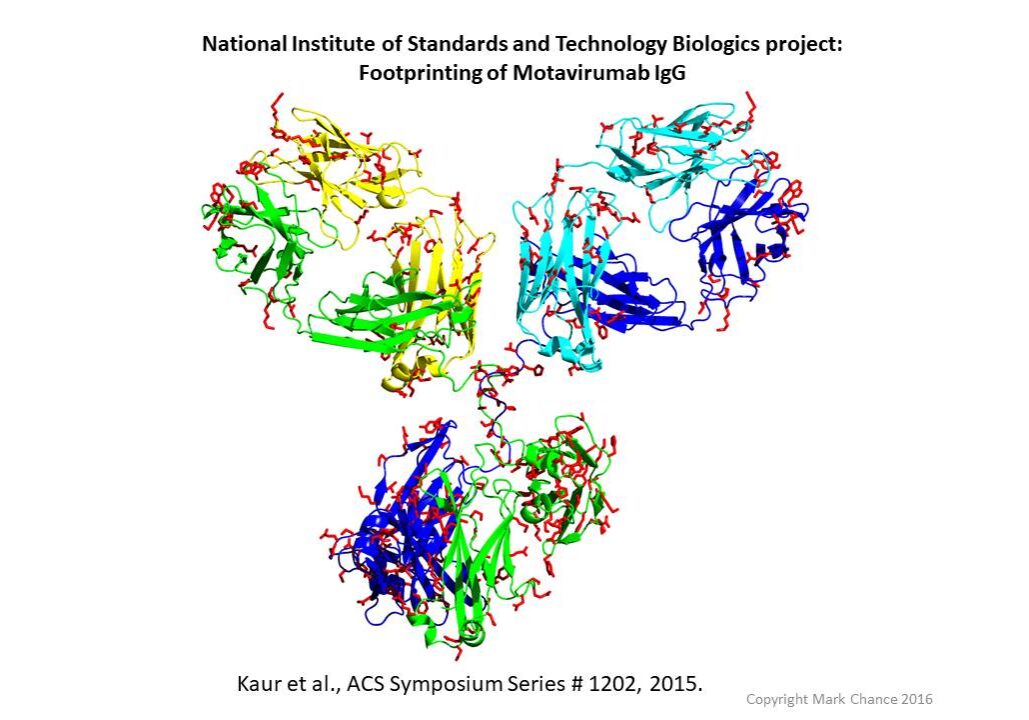
HRF antibody map. Red indicates labeled sidechains.
Contact us today to learn more about our protein footprinting technology.
